 Regulation and secretion of noncoding RNA by oncogenic RAS signaling
Regulation and secretion of noncoding RNA by oncogenic RAS signaling
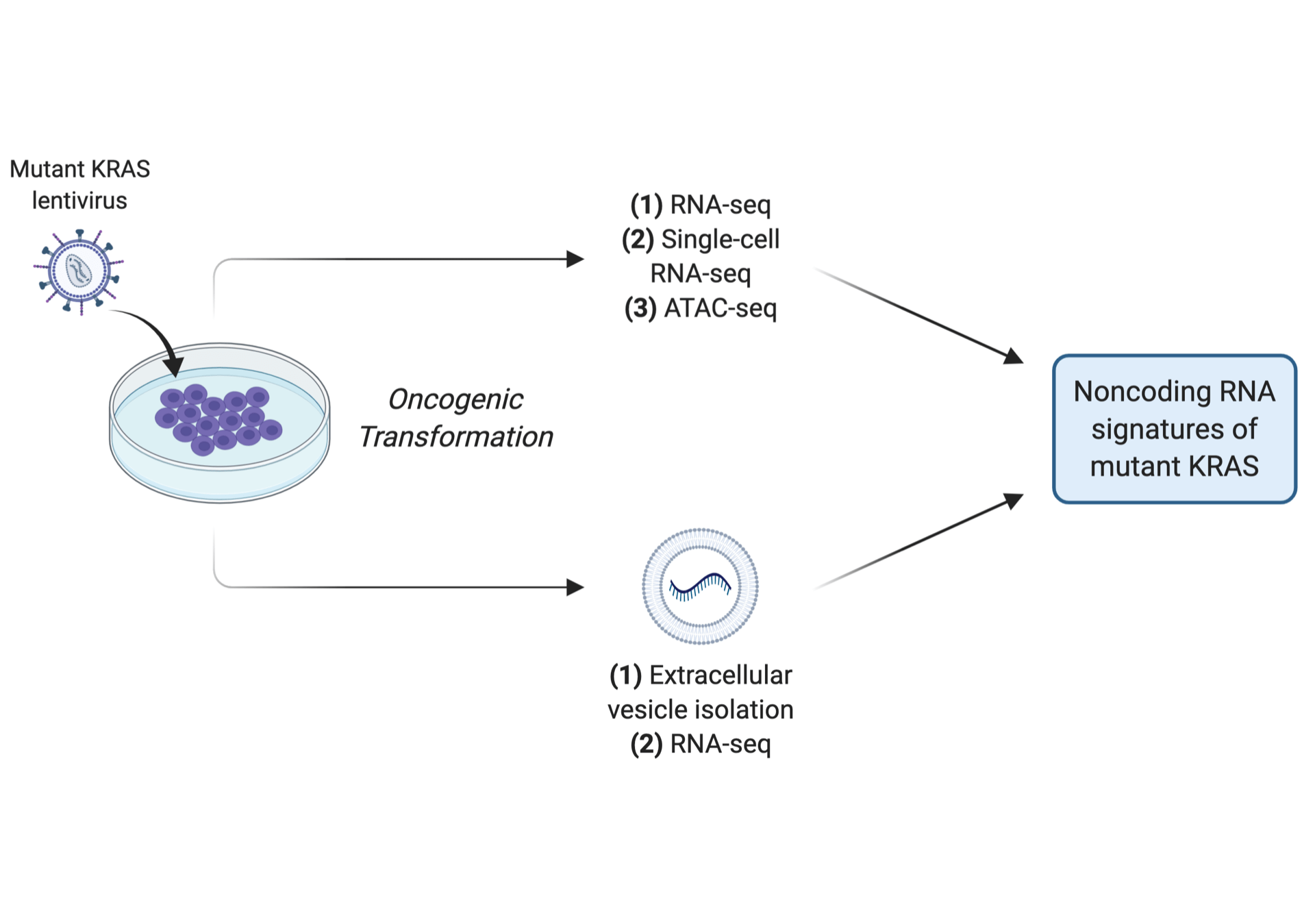
RAS genes are the most frequently mutated oncogenes in cancer. However, the effects of oncogenic RAS signaling on the noncoding transcriptome are unclear. We previously discovered that noncoding RNAs are coordinately regulated with RAS signaling genes in single cells during epigenomic reprogramming. To investigate how mutant KRAS regulates the noncoding transcriptome during the initial stages of cellular transformation, we characterize the composition of protein-coding RNA, long noncoding RNA, transposable-element derived repetitive noncoding RNA, and extracellular RNA, as well as global chromatin accessibility, using human lung or pancreatic cells that are transformed by constitutively active mutant KRAS. Our research aims to reveal the landscape of noncoding RNAs that are regulated by oncogenic RAS signaling during cancer initiation. (illustration created with BioRender.com)
Noncoding RNA biomarkers for precision health and cancer early detection
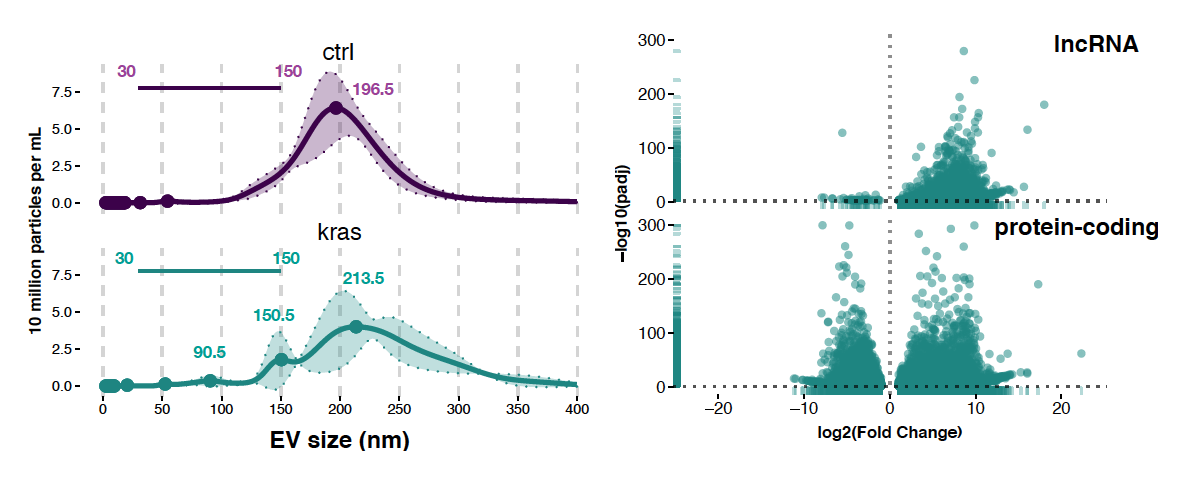
Noncoding RNAs that are released in extracellular vesicles (EVs), such as exosomes and microvesicles, are promising candidates as biomarkers for cancer early detection. We have found that in mutant KRAS cells, long noncoding RNAs (lncRNAs) are enriched in EVs relative to cells. To discover noncoding RNA biomarkers, we isolate EVs from cell culture and blood plasma and perform RNA sequencing. Our research aims to leverage these findings to develop non-invasive RNA liquid biopsy technology for the early detection of cancer and other diseases to enable precision health.